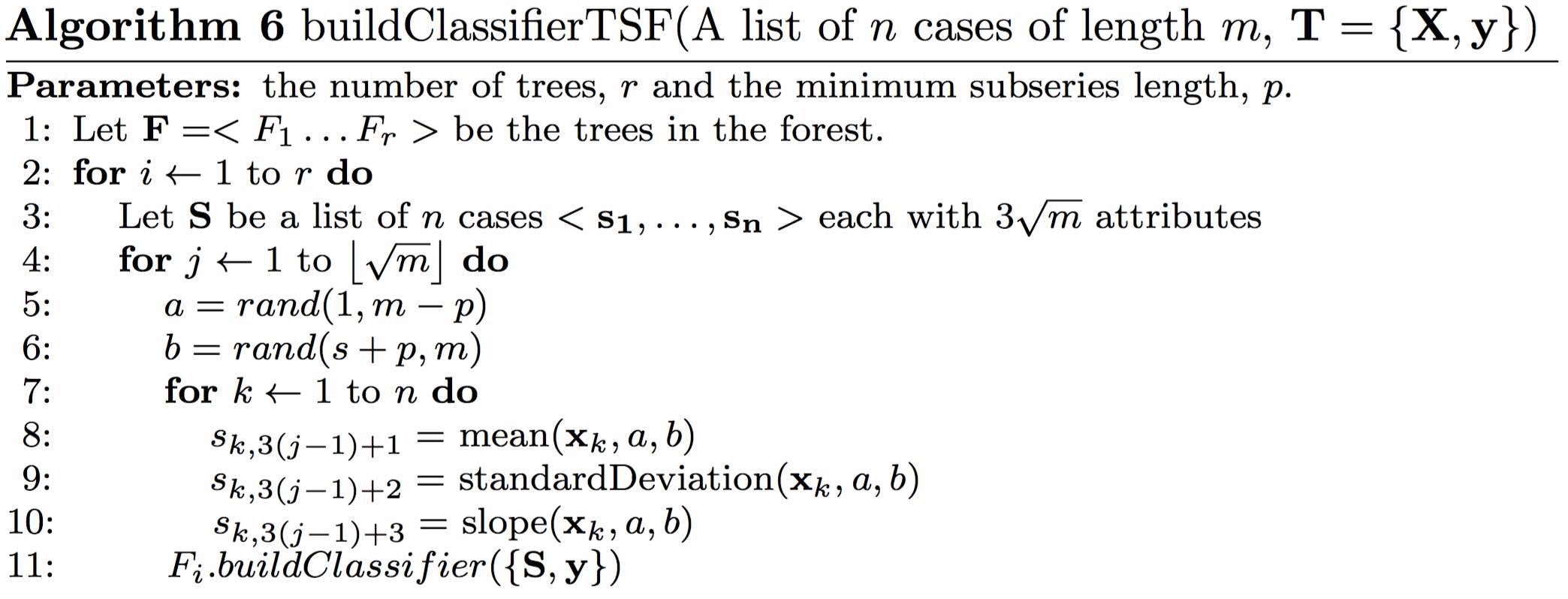
| Description: | Deng $et\:al.$ overcome the problem of the huge interval feature space by employing a random forest approach, using summary statistics of each interval as features. Training a single tree involves selecting $\sqrt{m}$ random intervals, generating the mean, standard deviation and slope of the random intervals for every series then creating and training a tree on the resulting $3\sqrt{m}$ features. Classification is by a majority vote of all the trees in the ensemble. The classification tree has two bespoke characteristics. Firstly, rather than evaluate all possible split points to find the best information gain, a fixed number of evaluation points is pre-defined. We assume this is an expedient to make the classifier faster, as it removes the need to sort the cases by each attribute value. Secondly, a refined splitting criteria to choose between features with equal information gain is introduced. This is defined as the distance between the splitting margin and the closest case. The intuition behind the idea is that if two splits have equal entropy gain, then the split that is furthest from the nearest case should be preferred. This measure would have no value if all possible intervals were evaluated because by definition the split points are taken as equi-distant between cases. We experimented with including these two features, but found the effect on accuracy was, if anything, negative. We found the computational overhead of evaluating all split points acceptable, hence we had no need to include the margin based tie breaker. We used the built in Weka RandomTree classifier (which is the basis for the Weka RandomForest classifier) with default parameters. This means there is no limit to the depth of the tree nor a minimum number of cases per leaf node. A more formal description is given in Algorithm 6. |